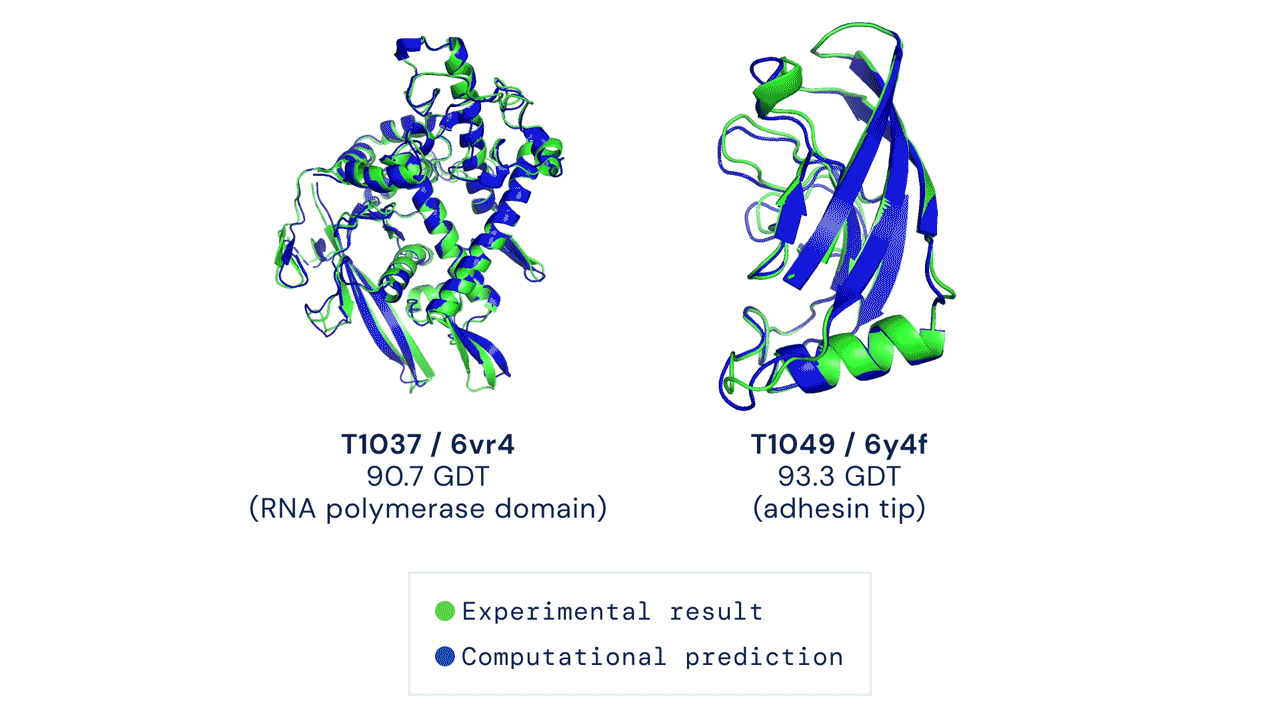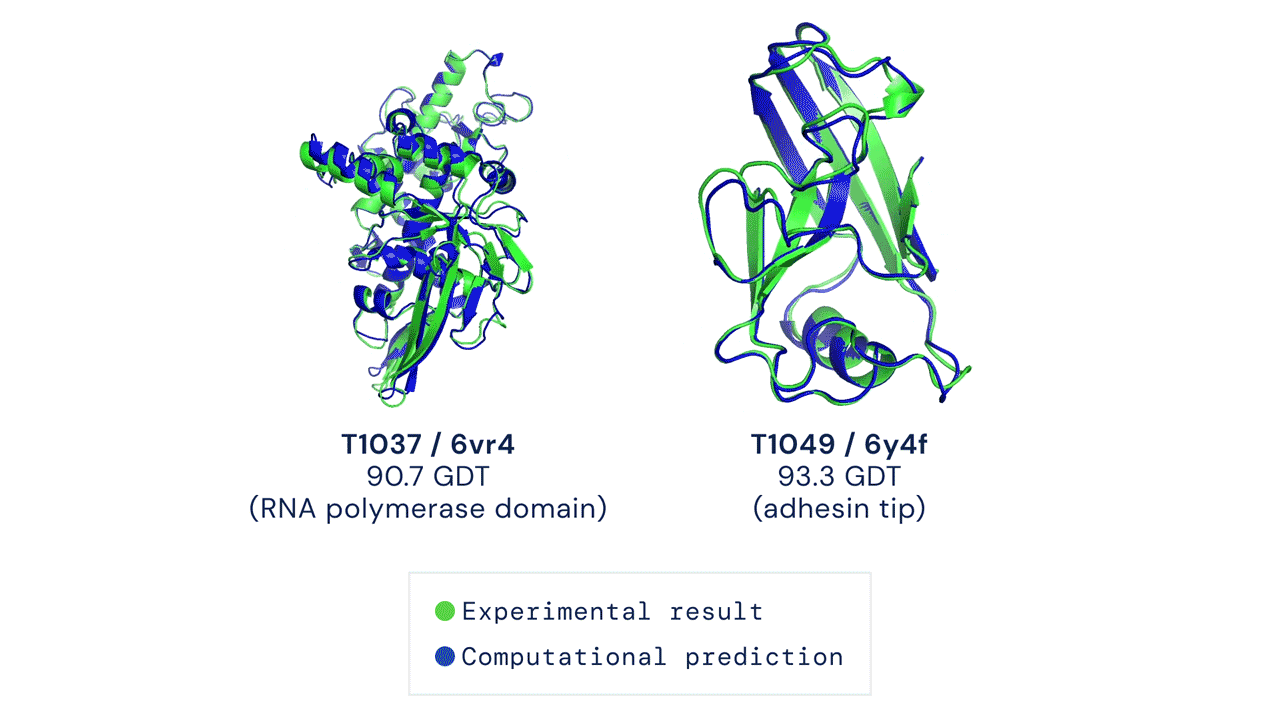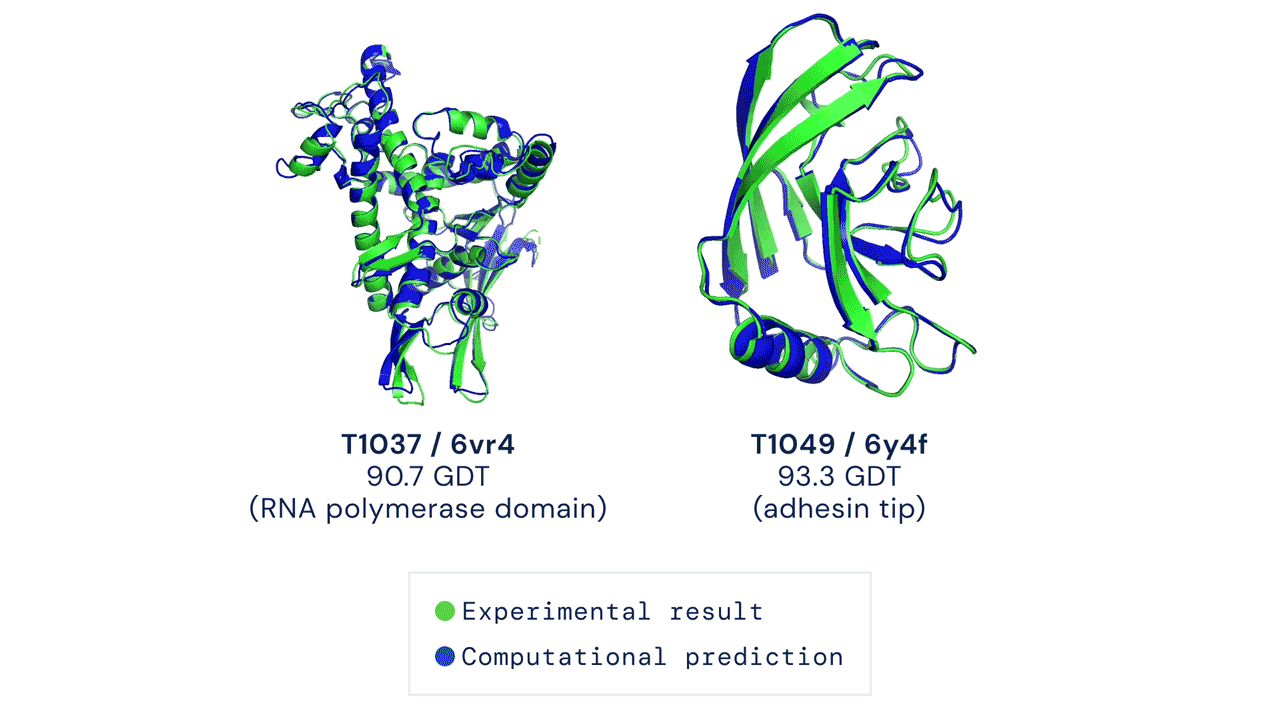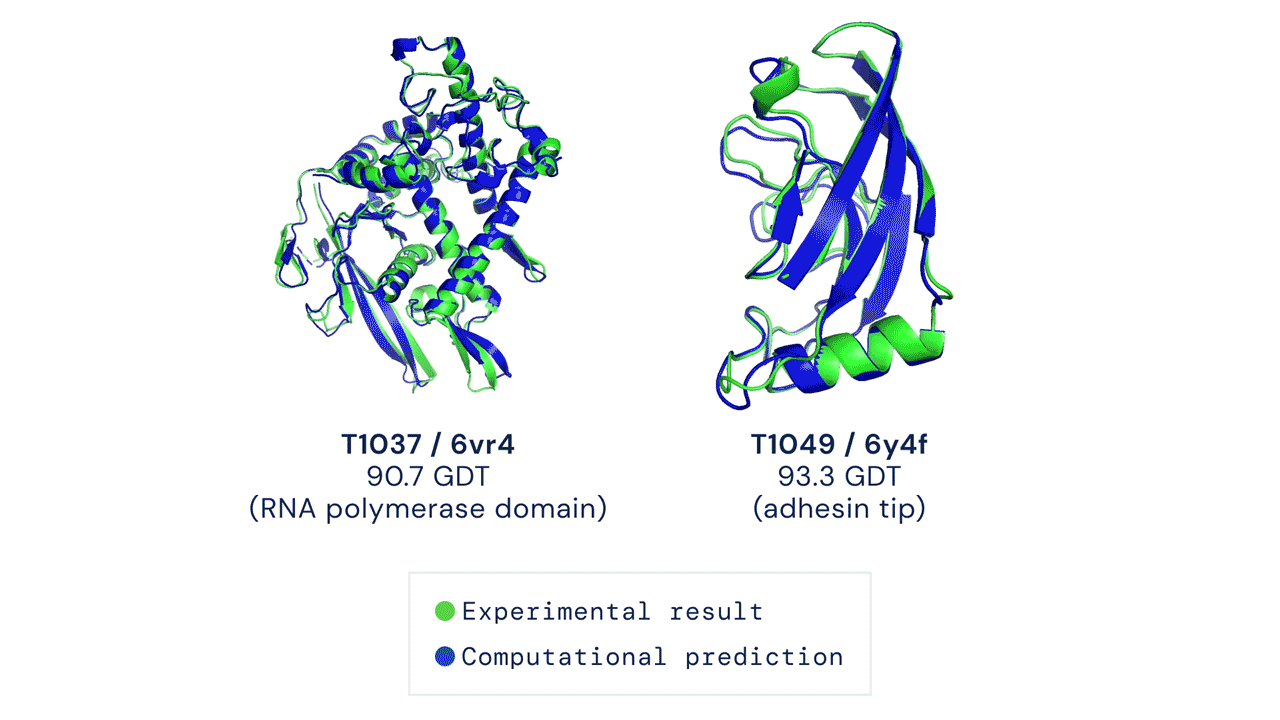
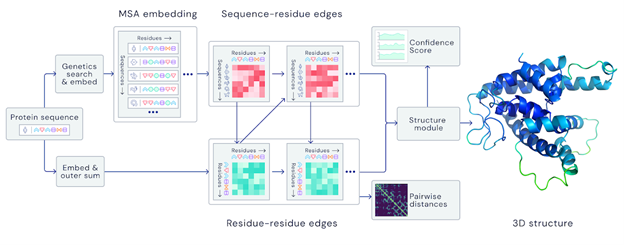
The Protein Folding Problem
Proteins are the building blocks of life. They are large complex molecules, made up of chains of amino acids, and what a protein does largely depends on its unique 3D structure. Figuring out what shapes proteins fold into is known as the “protein folding problem.” For decades and decades, one of biology’s biggest challenges has been finding a solution for the “protein folding problem” and is explained in the linked video below.
AI, DeepMind and Google Find Answers
Founded in 2010, DeepMind researches and builds safe AI (Artificial Intelligence) systems that learn how to solve problems and advance scientific discovery for all. They joined forces with Google in 2014 to accelerate their work. They’re a team of scientists, engineers, machine learning experts and more, working together to advance the state of the art in AI.
In a major scientific breakthrough, DeepMind’s AI system AlphaFold has been recognized as a solution to this grandest of all biological problems – the “protein folding problem.” Here is an excellent video explaining AlphaFold and the making of a scientific breakthrough.
According to Professor Venki Ramakrishman, Nobel laureate and President of the Royal Society,
This computational work represents a stunning advance on the protein-folding problem, a 50-year-old grand challenge in biology. It has occurred decades before many people in the field would have predicted. It will be exciting to see the many ways in which it will fundamentally change biological research.
Potential Impact for Amyloidosis
For diseases which originate with misfolded proteins, such as amyloidosis, “investigators have been doing this exercise by ‘brute force’ until now,” according to Dr. Angela Dispenzieri from the Mayo Clinic. This AI research is likely to open a whole new world of insight and answers, from which new and more effective treatments can be developed.
Marina Ramirez-Alvarado, Ph.D., whose research laboratory at the Mayo Clinic studies misfolding and amyloid formation in light chain amyloidosis, had this to say.
The protein folding problem, one of the most important scientific questions of the 20th century is making headlines today with the artificial intelligence work from DeepMind. It is clear that DeepMind will provide important basic understanding of the folding process and will significantly benefit those amyloidosis diseases that involve secreted, folded proteins, such as light chain (AL), and Transthyretin (ATTR) amyloidosis.
Dr. Morie Gertz, a hematologist/oncologist from the Mayo Clinic who has decades of clinical experience with amyloidosis, weighs in on some of the possible outcomes from this ground-breaking research.
The ability to predict protein folding in three dimensions may result in the ability to predict which protein sequences are likely to form amyloid fibrils. In light chain amyloidosis this could allow for long-term monitoring of selected patients likely to develop amyloidosis. This would permit extremely early diagnosis long before symptoms developed. It would also allow for the exploration of why wild-type TTR amyloidosis forms amyloid fibrils in the heart in some patients but not in others.
However, it won’t answer all questions …
Dr. Vaishali Sanchorawala, director of Boston University’s Amyloidosis Center offers these words of perspective.
The “protein folding problem” that DeepMind’s AlphaFold is designed to solve is predicting the native, functional state of a protein from just its amino acid sequence. Amyloidosis, though, is caused by our bodies’ failure to solve that problem, resulting in misfolded and aggregated proteins. AlphaFold’s remarkable achievement can definitely help to better understand native structure of amyloidogenic light chain proteins. However, amyloid fibrils are different from the native states of their precursor proteins and therefore the adaptation of AlphaFold to study protein misfolding and aggregation, perhaps by predicting the structures of complex amyloid fibrils, might be better able to predict the effects of mutations that alter people’s risk of developing amyloidosis.
In closing …
AI is rapidly advancing the knowledge of protein misfolding, unlocking answers for amyloidosis which should lead to earlier diagnosis, improved treatment, and better patient survival.
———————————————————-
Sources:
Morie A. Gertz, M.D., M.A.C.P.
Marina Ramirez-Alvarado, Ph.D.
High Accuracy Protein Structure Prediction Using Deep Learning
John Jumper, Richard Evans, Alexander Pritzel, Tim Green, Michael Figurnov, Kathryn Tunyasuvunakool, Olaf Ronneberger, Russ Bates, Augustin Žídek, Alex Bridgland, Clemens Meyer, Simon A A Kohl, Anna Potapenko, Andrew J Ballard, Andrew Cowie, Bernardino Romera-Paredes, Stanislav Nikolov, Rishub Jain, Jonas Adler, Trevor Back, Stig Petersen, David Reiman, Martin Steinegger, Michalina Pacholska, David Silver, Oriol Vinyals, Andrew W Senior, Koray Kavukcuoglu, Pushmeet Kohli, Demis Hassabis.
In Fourteenth Critical Assessment of Techniques for Protein Structure Prediction (Abstract Book), 30 November – 4 December 2020. Retrieved from here.